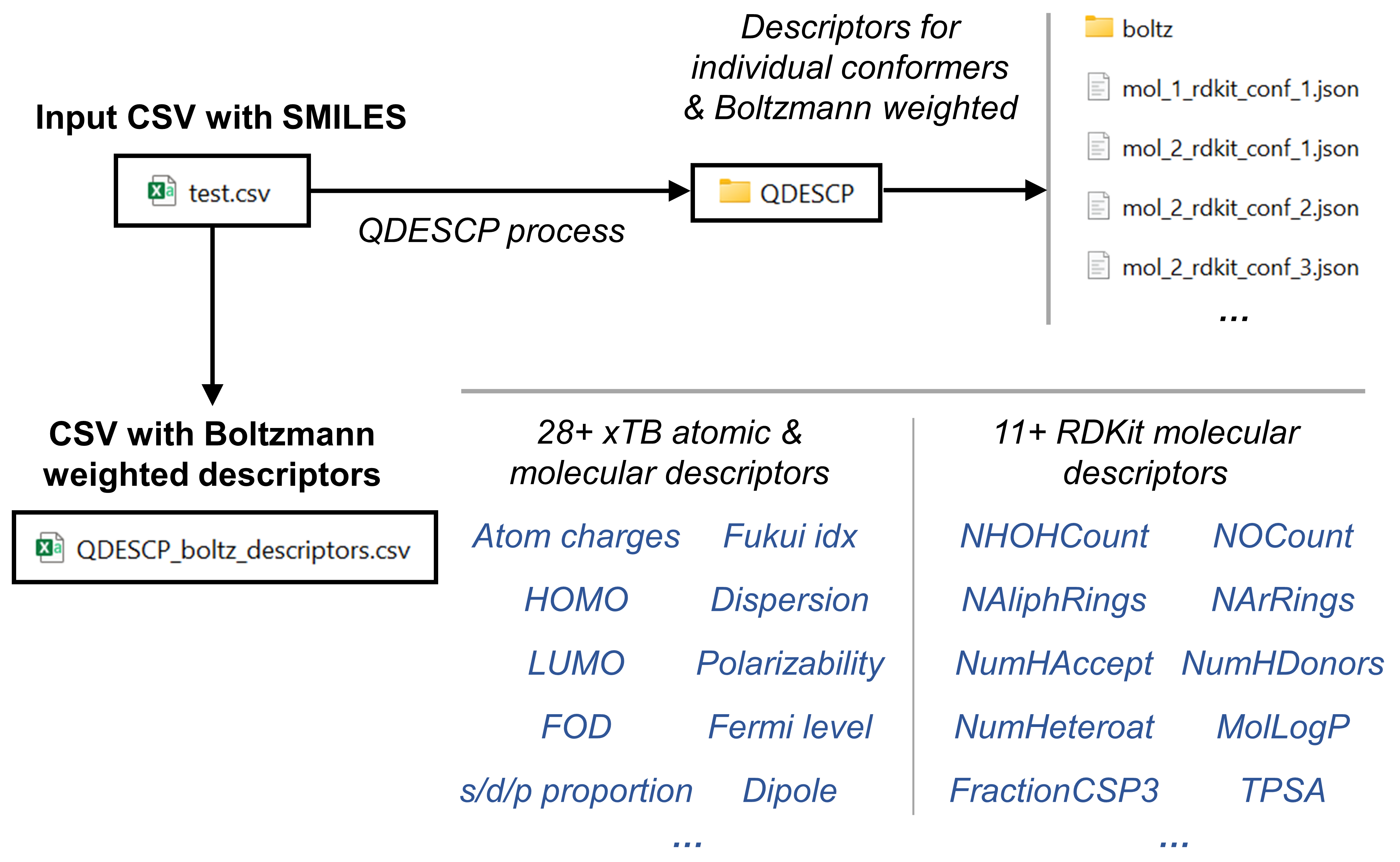
Descriptor Generation
In this example we are going to generate a collection of xtb-derived chemical descriptors as well as a collection of RDKit-derived descriptors. We are going to store them in .json format for each molecule. And we are going to create a csv file with the boltzmann averaged values of the descriptors that we have calculated per each molecule. The following scheme summarizes the contents of this example.
We are starting from a 'test.csv' file containing the SMILES of the molecules whose chemical descriptors we are going to calculate:
SMILES,code_name
CN1[N]C=NN(C)C1=O,mol_1
CC1(C)N([O])[CH]N([O])C1(C)C,mol_2
CC1(C)CCC(C)(C)N1[O],mol_3
CC1(C)C=CC(C)(C)N1[O],mol_4
CC1(C)CCCC(C)(C)[N+]1[O-],mol_5
In this case we are going to start by generating some conformers of these molecules using rdkit (for more details on the conformer generation please check the Conformer Search section).
from aqme.csearch import csearch
csearch(input='test.csv',
program='rdkit')
Next we proceed to generate the descriptors which is fully automated by the QDESCP module.
from pathlib import Path
from aqme.qdescp import qdescp
conformer_files = [str(filepath) for filepath in Path('CSEARCH').glob('*.sdf')]
qdescp(files=conformer_files,
program='xtb',
boltz=True)